 |
 |
| Download |
Download |
| OSX 10.11+ (M1 Universal) |
|
| Click the icons above to download the latest ApE (v3.1.8, July 10, 2025) |
| A list of updates and bug fixes. |
| Slides from a series of presentations describing some of the features of ApE |
I've started a YouTube channel with tutorial videos. |
| |
| |
Download demo theme files for circular graphic maps. |
New installs will open in inline feature mode (showing the bottom stand and features in the sequence window). If you are editing large sequences or you prefer the compact view, you can turn off inline mode in Preferences (aka Settings). |
| Click here to download the latest ApE for Linux. |
Some Windows systems will need to temporarily disable Windows Defender to install.
|
Click here  to make a voluntary donation in support of ApE. to make a voluntary donation in support of ApE. |
|
|
- Runs in Windows (XP, Vista, 7, 8, and 10) and Mac (OS X v10.11 and above)
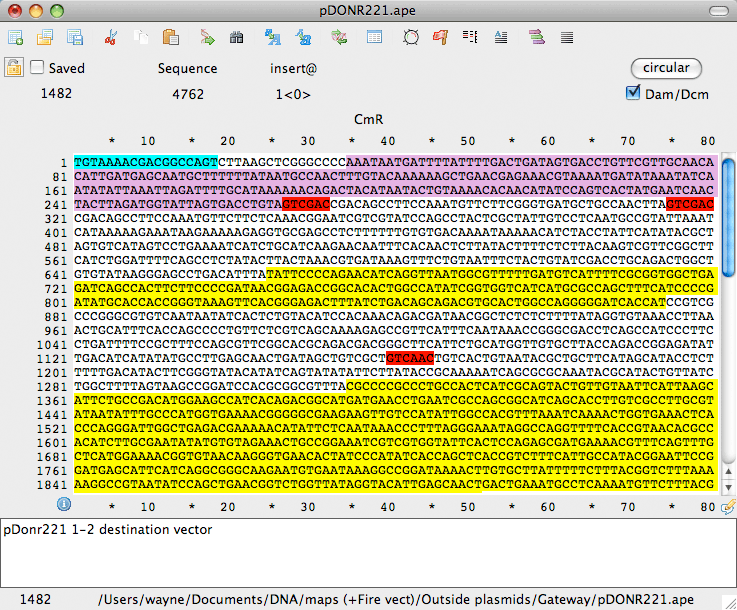
- Highlights restriction sites in the editing window
- Accurately reflects Dam/Dcm blocking of enzyme sites
- Highlights text using pre-defined and custom feature libraries
- Shows translation, Tm, %GC, ORF of selected DNA in real-time
- Reads DNA Strider, Fasta, Genbank and EMBL files
- Saves files as DNA Strider-compatible or Genbank file format
- Highlights and draws graphic maps using feature annotations from genbank and embl files
- Directly BLASTs selected sequence at NCBI or wormbase
|
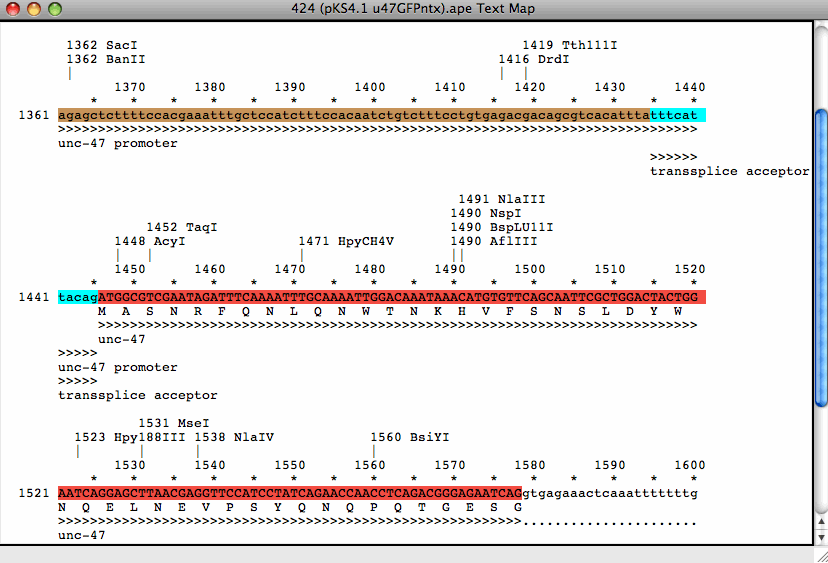
- Text map shows DNA sequence, translation, and features as text-based graphics
| 
|
|
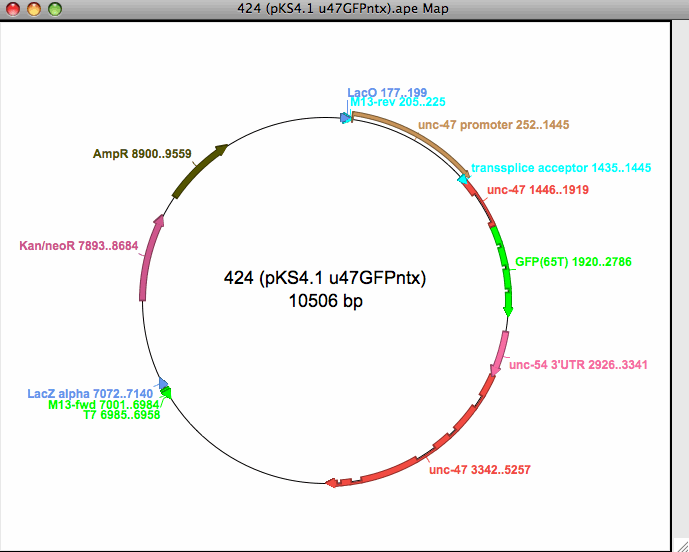
- Creates graphic restriction maps- linear or circular with features indicated
- Connects graphic and text features with hyperlink double click
- Saves graphics as encapsulated postscript or scalable vector graphics
- Copy and save graphics as Windows metafiles (MS Windows only)
|
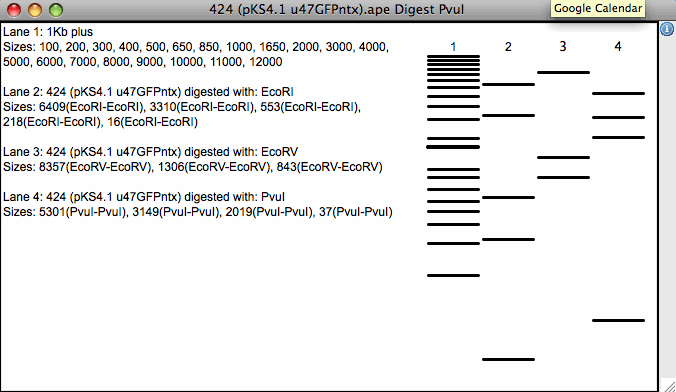
- Virtual restriction digest
- Draws pre-defined and user-defined DNA ladders
- Connects bands to text by double-click
| 
|
|
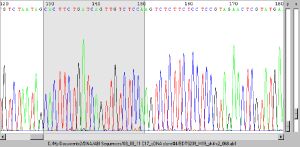
- Reads ABI sequencing trace files
- Sequences in ABI traces can be aligned directly to a reference sequence, with the alignment hyperlinked back to te trace.
|
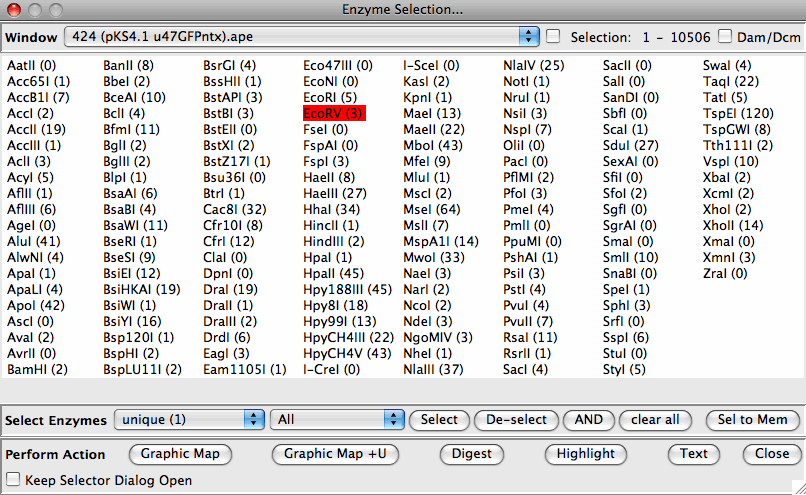
- Selects sites matching multiple criteria (union/intersection- cut frequency, site type) in all open windows
- Selects sites that cut more often in one sequence than another (for snip-SNP detection or diagnostic digests)
- Has user defined enzyme grouping to distiguish eg. enzymes currently in stock.
- Allows users to define new enzymes by name and recognition site
- Imports DNA Strider format files (simple enzyme, site lists) available from REBASE
| 
|
Other Features:
|
- Most analysis windows are hyperlinked to their corresponding sequences, including:
- Graphic Maps
- Text maps
- Virtual Digests
- Alignments (including ABI sequences)
- Silent Sites
- Translation
- Primer Find
- Uses custom feature definition libraries, which allow:
- Quick annotation of sequence
- Quick searching and highlighting of all available primers that you (or others) have that hybridize to a sequence
- Sequence to be annotated and visualized in multiple ways quickly and efficiently
- Graphic maps that show primer binding sites and all interesting sequence features
- Translates sequences with optional DNA alignment
- Finds potential primers matching user criteria (length, Tm, %GC, self/other complementarity)
- Aligns two DNA sequences (or any combination of sequence and ABI trace), with the alignment hyperlinked to the original sequence
- Finds translationally silent restriction sites
- Draws graphic ORF maps
|
Click here

to make a voluntary donation in support of ApE.
You can now export genomic regions from Wormbase directly: In the upper right drop-down menu button, select "Download Track Data". Click the "Configure..." button. Dump selected features using version 3 Across currently visible region. Check "Save to File". Check "Embed DNA sequence". UNCHECK "Include track configuration data". Click "Go". Open the resulting .gff file in the latest ApE.
Alternatively, you can export a genomic region (from the genome viewer) as a FASTA formatted file (using the menu on the upper left). You can add the feature tracks by downloading the GFF3 feature track files using the same menu. In ApE, open the FASTA file, then use the Features menu to open the GFF3 track info.
Another way to go is to take the gene model (from a gene page), paste it into an ApE window and then select all, make a new feature (Feature menu), and in the edit feature window that appears press the "upper case only" button.
If you think that ApE doesn't find all of the ClaI sites (or XbaI or BclI) that you KNOW are present in your sequence, turn off the Dam/Dcm methylation on your sequence and try again.
See here for more information.
Click here  to make a voluntary donation in support of ApE.
to make a voluntary donation in support of ApE.
Last modified: July 10, 2025


 to make a voluntary donation in support of ApE.
to make a voluntary donation in support of ApE.





 to make a voluntary donation in support of ApE.
to make a voluntary donation in support of ApE.
 to make a voluntary donation in support of ApE.
to make a voluntary donation in support of ApE.