Golden Gate Reactions
Golden Gate Assembler
The Golden Gate assembler is a tool for assembling DNA fragments from pre-designed libraries of compatible parts.
There are several widely used collections, like C. elegans SapTrap, MoClo, etc.
For this example, we will assemble a four-part SapI-based Golden Gate reaction.
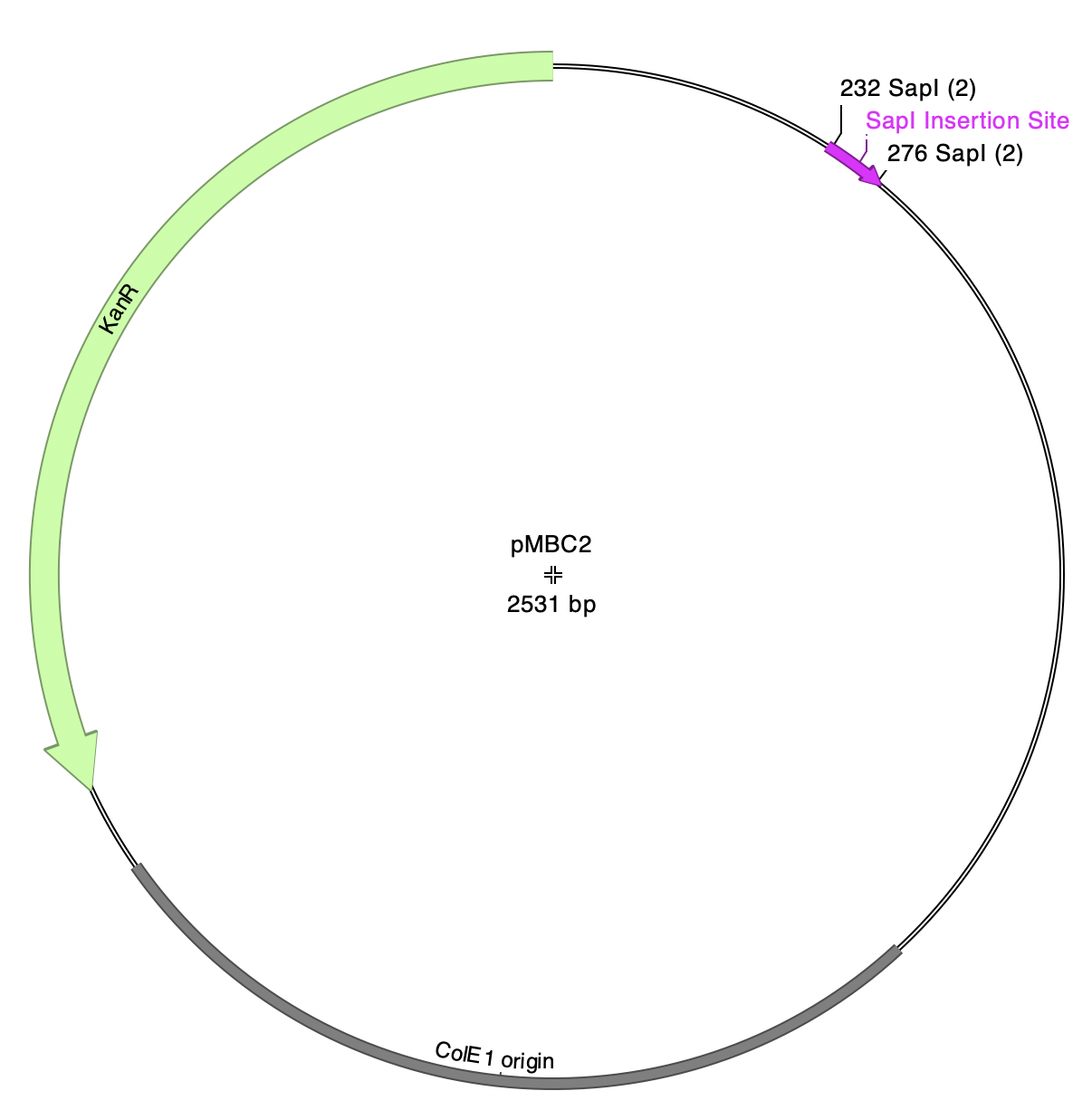

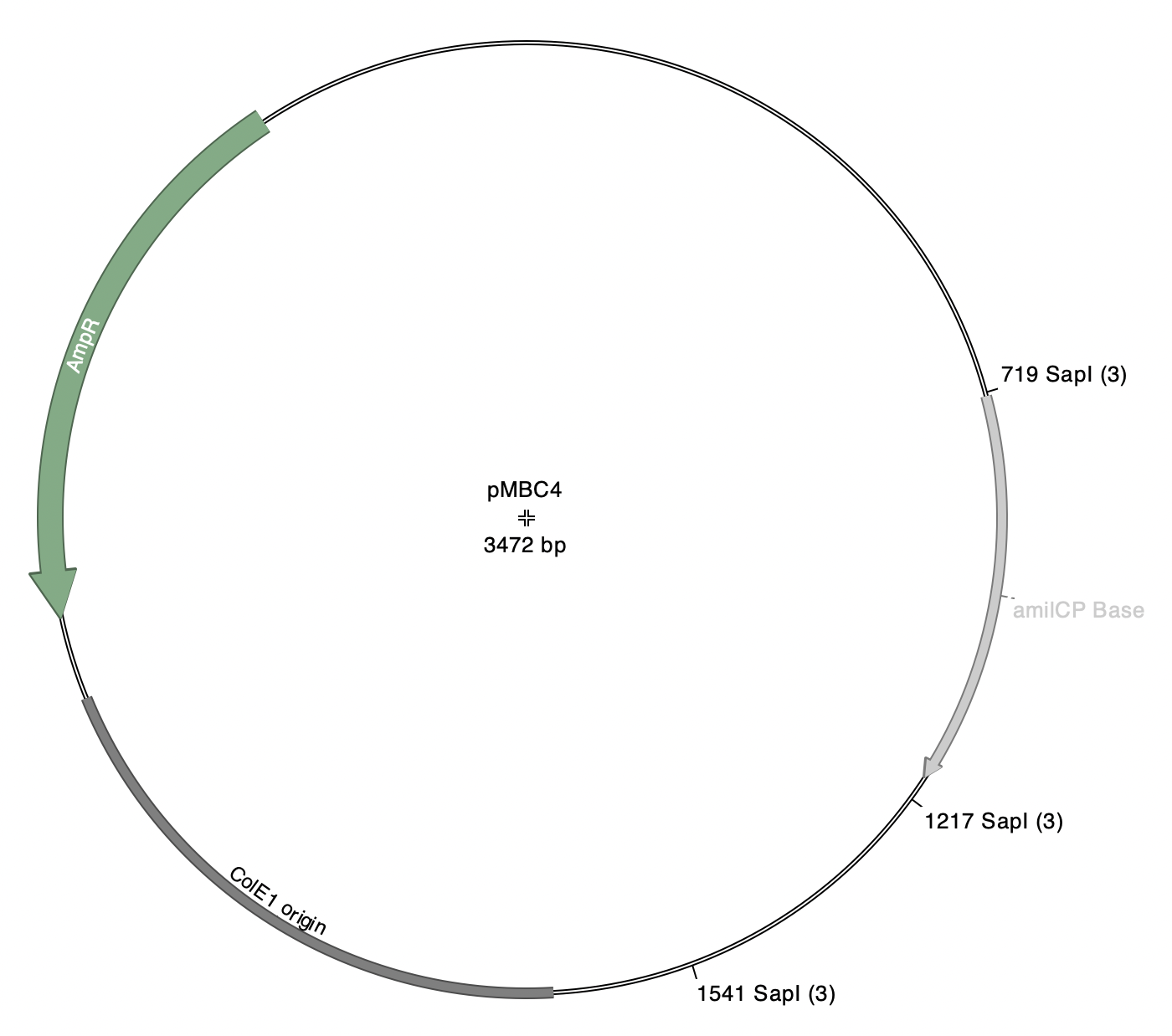
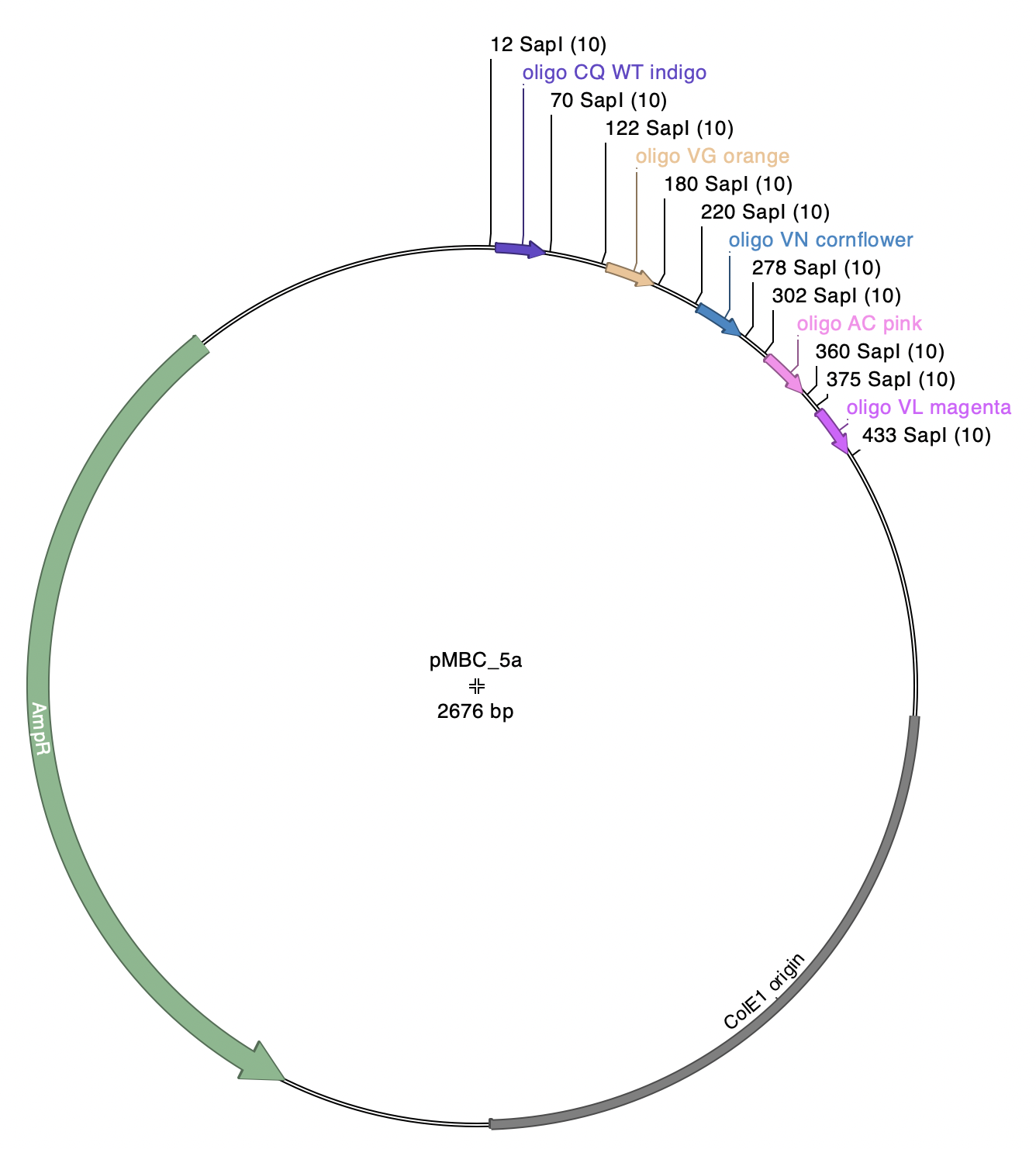
Note: pMBC5a has five SapI-flanked fragments. Each of these fragments has identical SapI overhangs (atg-cgt), so there are five independent product plasmids that can be generated by this reaction.

pMBC2 has outward-facing SapI sites and is the plasmid destination for this reaction:
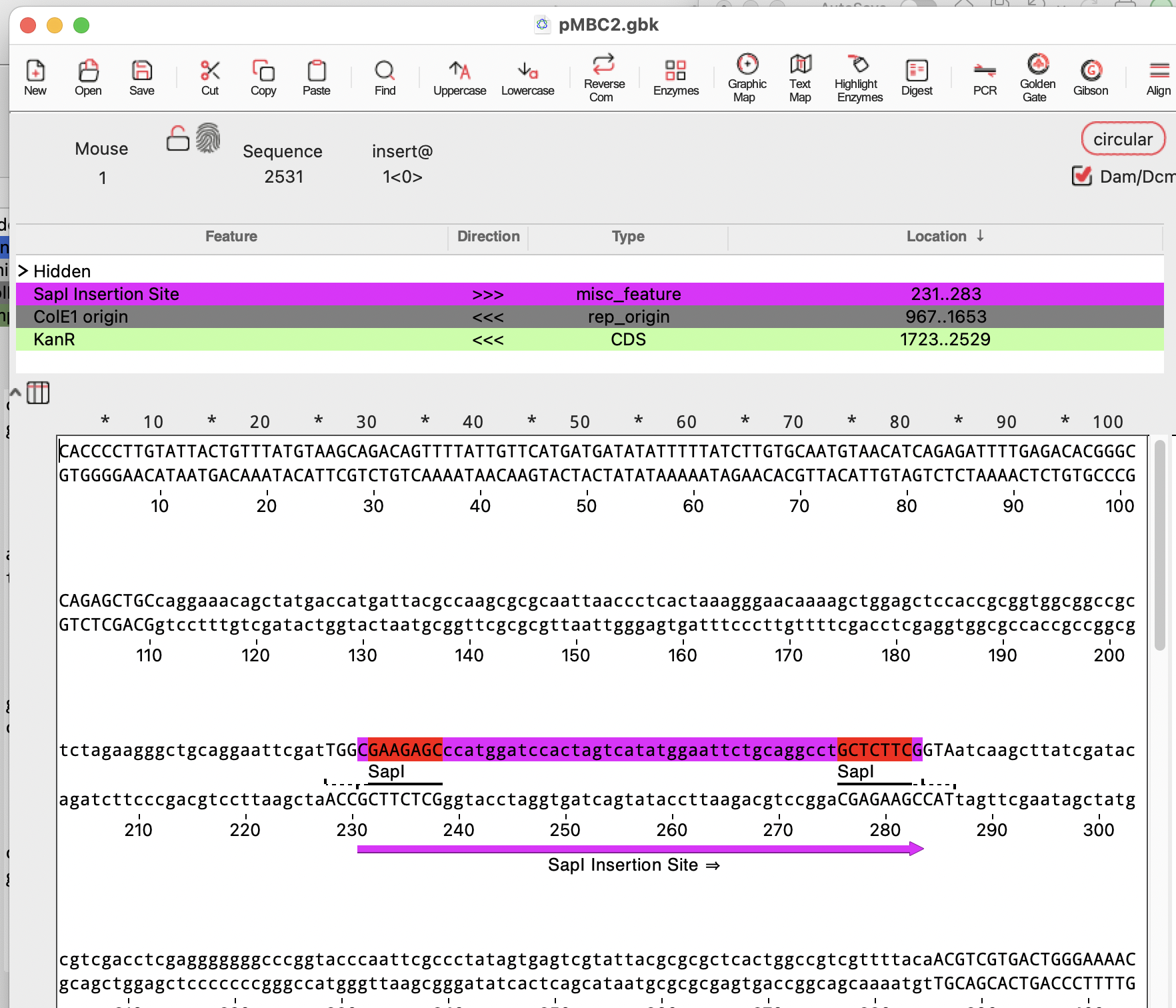
With these sequences open, Select the Golden Gate assembler tool:
The Assembler tool dialog will open:
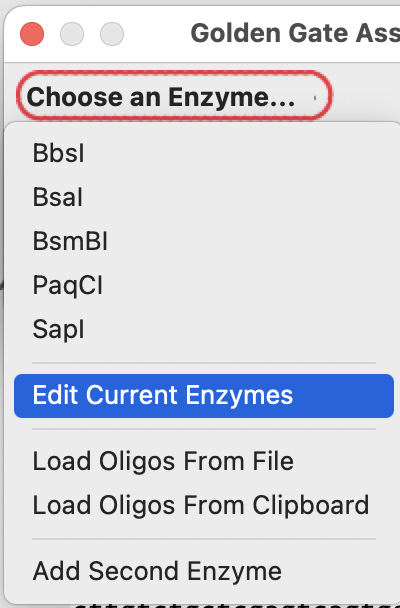
Press the “Choose an Enzyme” menu button:
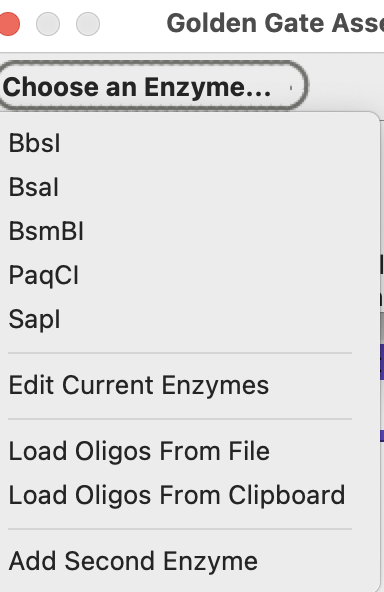
If SapI isn’t in your current enzyme set, choose “Edit Current Enzymes”:
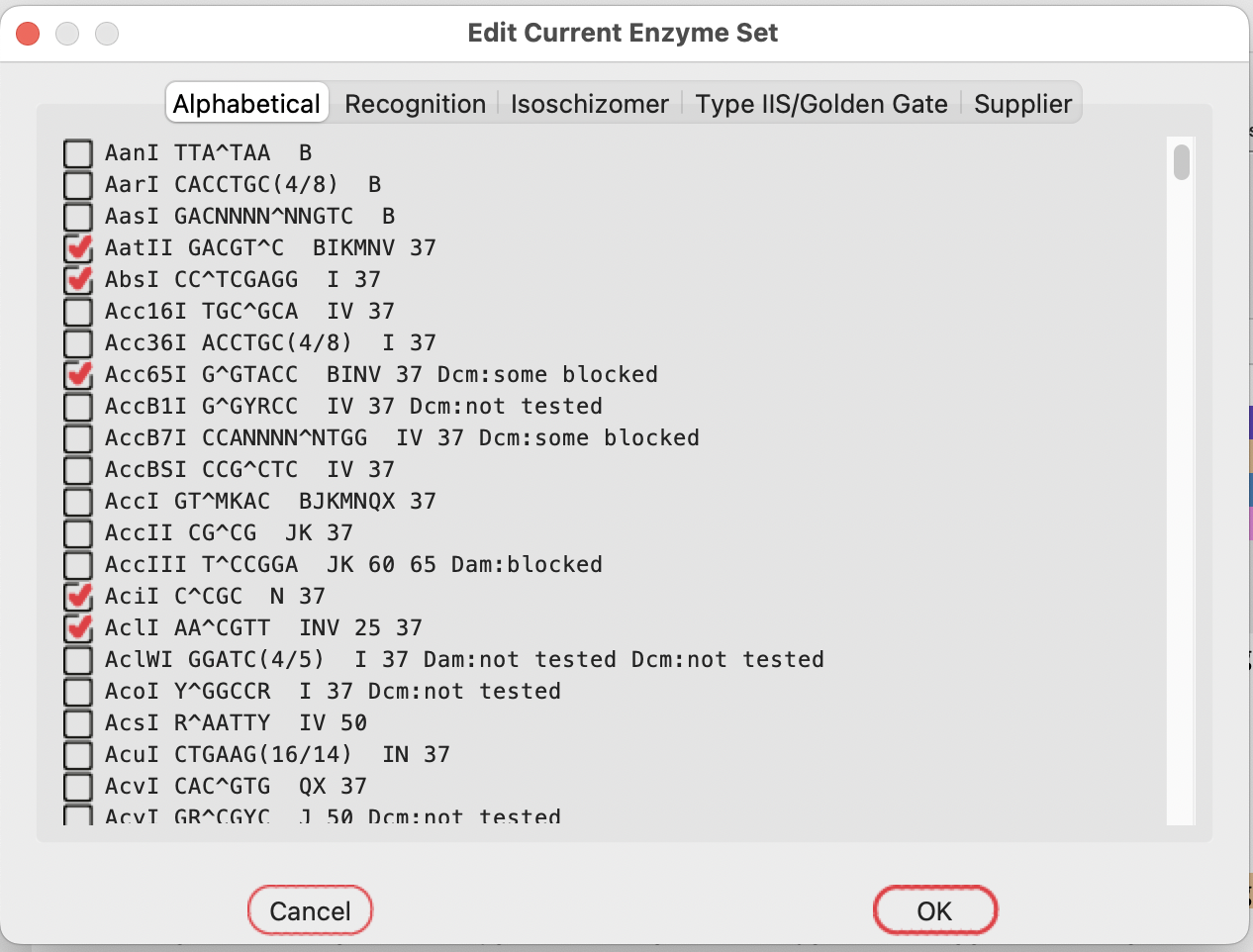
Select the TypeIIS tab:
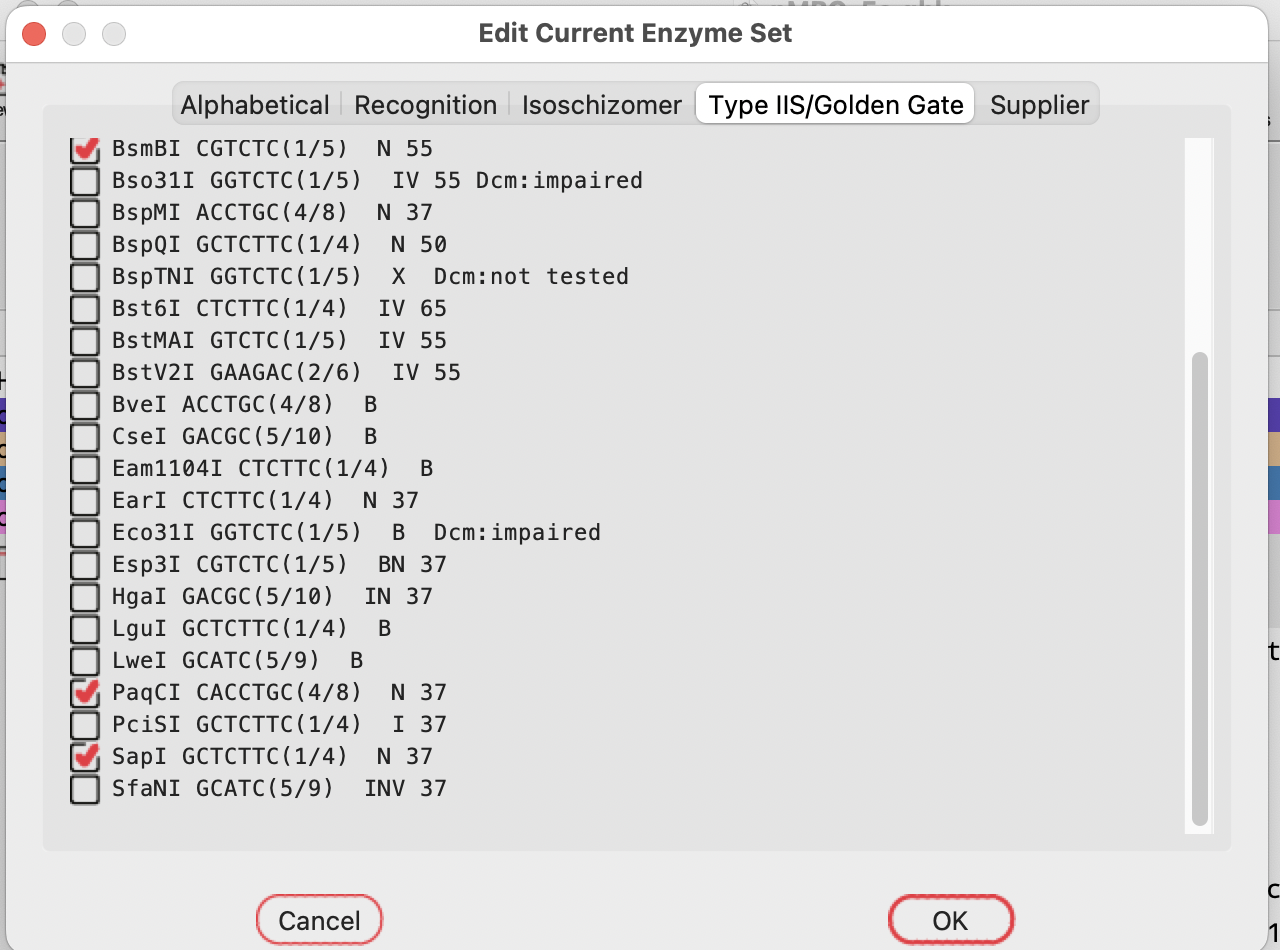
Then select SapI to add that enzyme to your current set:

Once the enzyme is added, you will be asked to save the current default enzyme set when you quit the ApE program.
If you chose to save the enzyme set, you will not have to re-add the SapI enzyme the next time you use the Golden Gate Assembler tool.
Return to the Golden Gate assembler tool and select SapI:
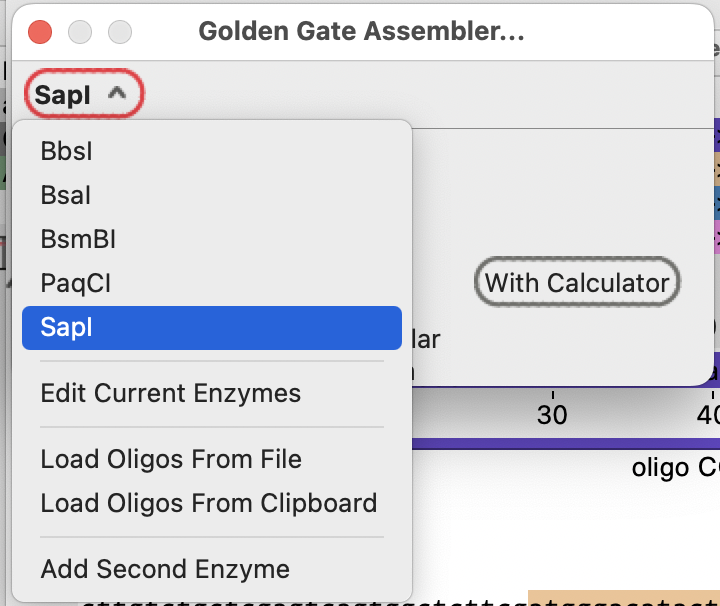
If there are SapI-flanked fragments in the currently open sequences, the menu button changes to “Choose a Fragment”
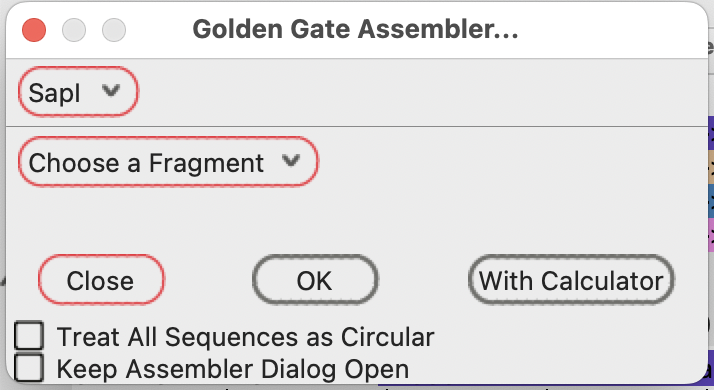
You can select any of the SapI-flanked fragments in the drop down menu:
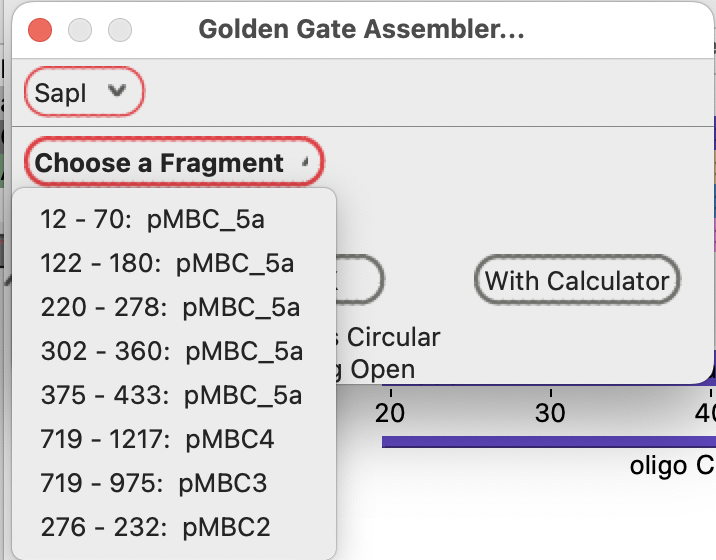
Selecting pMBC2 as the first fragment fills in the subsequent compatible fragments:
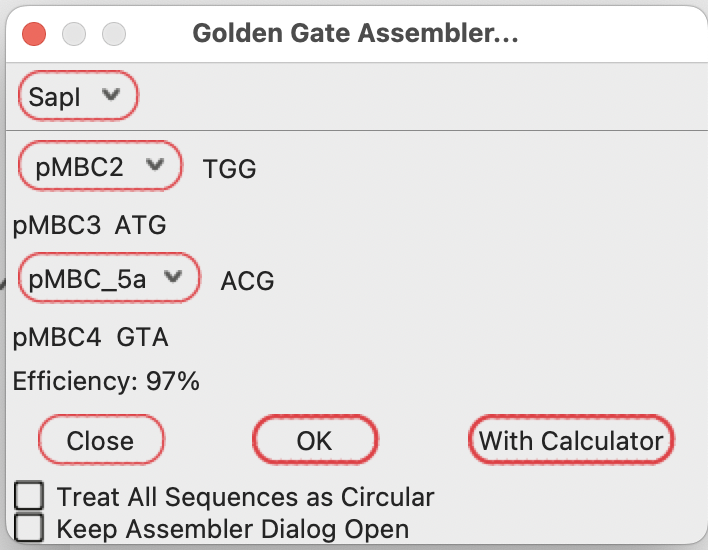
The three-base overhang at the right end of each fragment is shown to the right of each fragment.
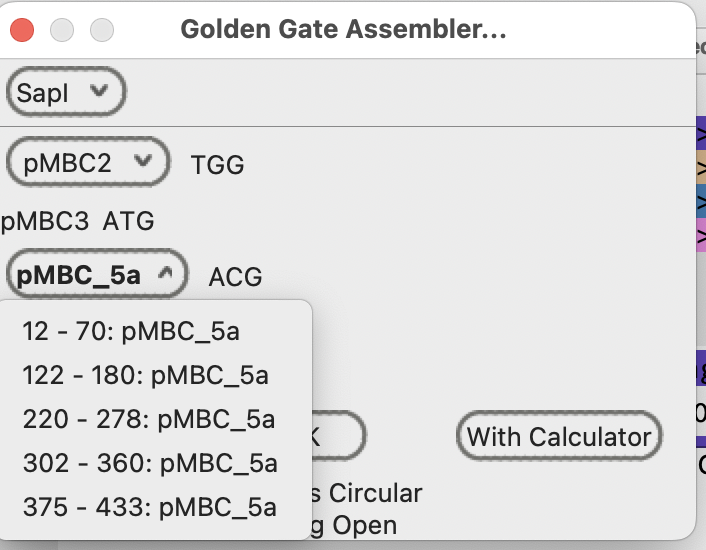
Note that there are multiple choices for the pMBC5a fragment position.
They are noted in the drop down menu by their sequence position number in the plasmid sequence.
The efficiency is calculated as on target vs. off target overhang ligation as measured in Pryor et al. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0238592
You can generate the Golden Gate reaction product by pressing “OK”.
If your sequences are saved as linear sequences, but they are actually circular, you can either circularize the sequences (the preferred option) or press the “Treat all sequences as Circular” check button
If you would like to assemble multiple different products in a single session, select the “Keep Assembler Dialog Open” check button:
The dilaog will stay open after you press the “OK” button to generate a new sequence window with the calculated Golden Gate product.
If you will be using DNA olios that have been annealed together to produce single stranded overhangs like this:
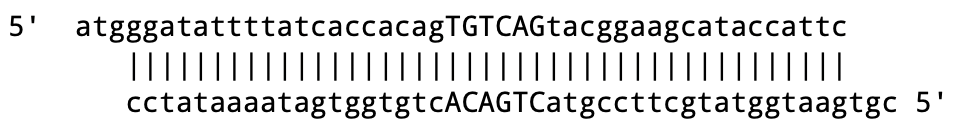
Copy the single stranded oligo sequences from a text editor (or spreadsheet columns) in 5’ to 3’ normal sequence notation:

In the Golden Gate Assembler dialog you can select “Load Oligos from Clipboard” to have the dialog consider the possible double stranded fragments that could be generated from the single stranded sequences. You can also save the sequences to a text file and load the sequences from that file using the “Load Oligos From File” command.
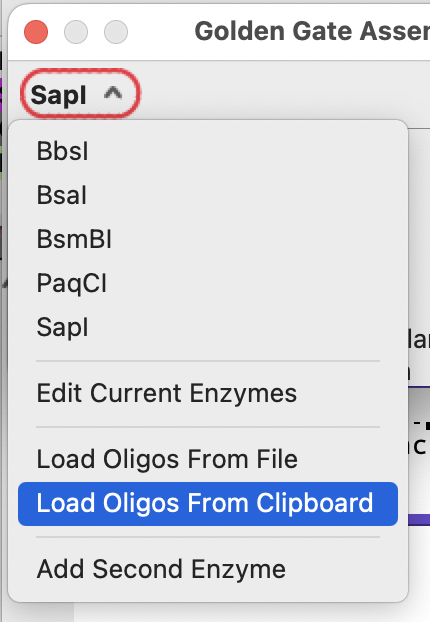
The double stranded oligos will then be available from the fragment drop down menu:

And will be available as an intermediate fragment in an assembly:
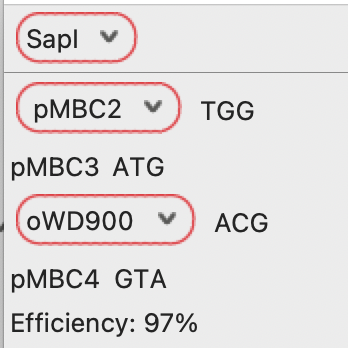
Pressing “With Calculator” will generate the same reaction product, but will also auto-populate the Molecular Reaction Calculator with the input fragment information:
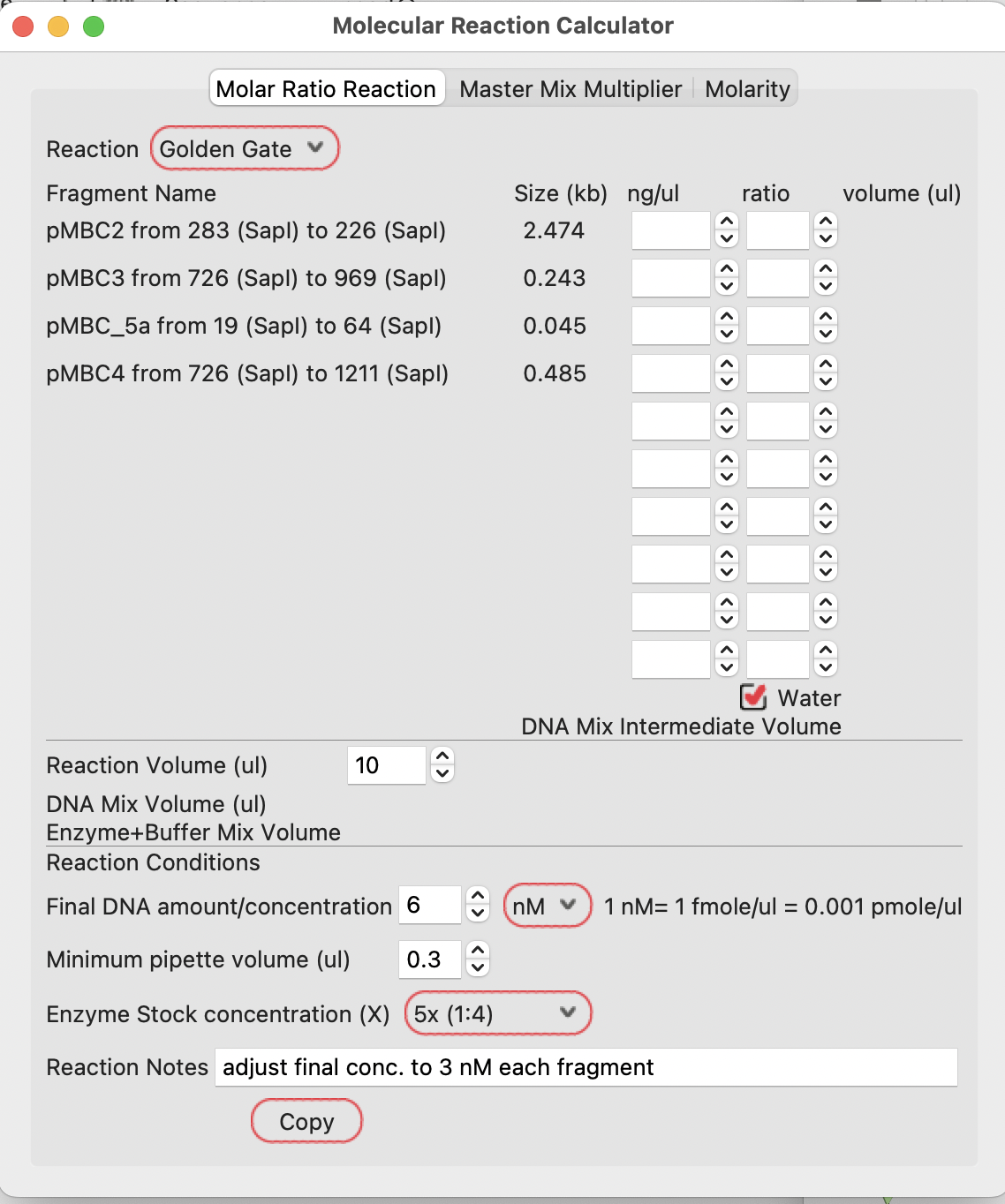
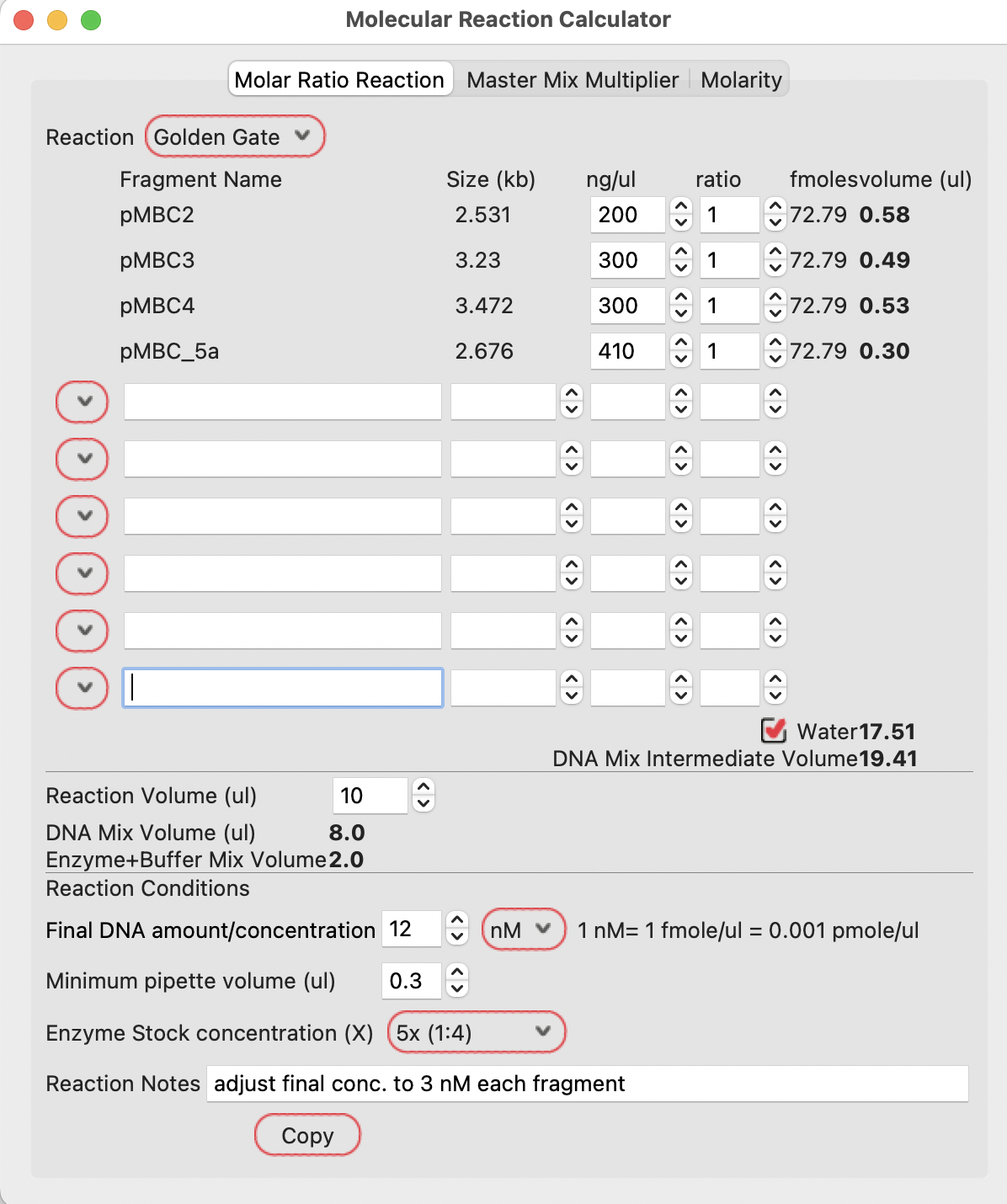
If you enter the concentration of each of your fragments and your preferred molar ratio in the reaction, it will calculate the required volumes in the Golden Gate DNA mix:
**Note that this calculation is based on each fragment in isolation (as if each is a PCR product, for example).
If the fragments are contained in a larger piece of DNA (a plasmid, for example), you need to reset the molecular calculator values:
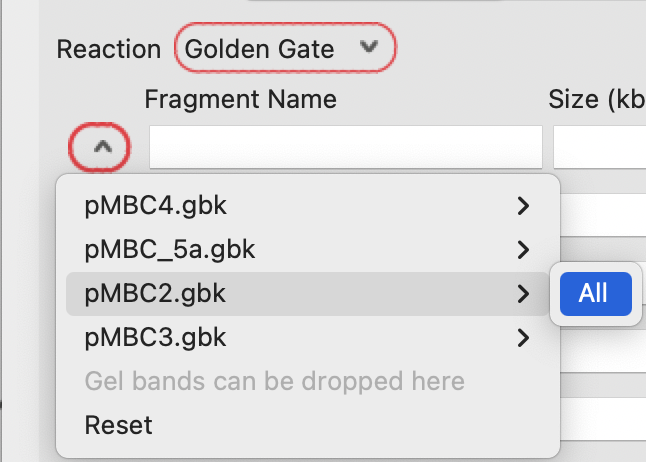
Then select the full plasmid sequences individually
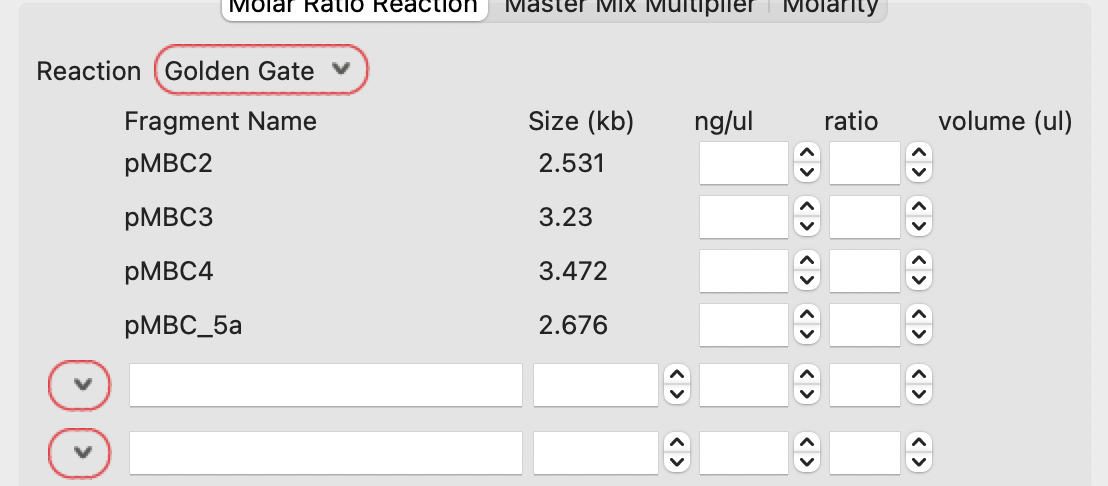
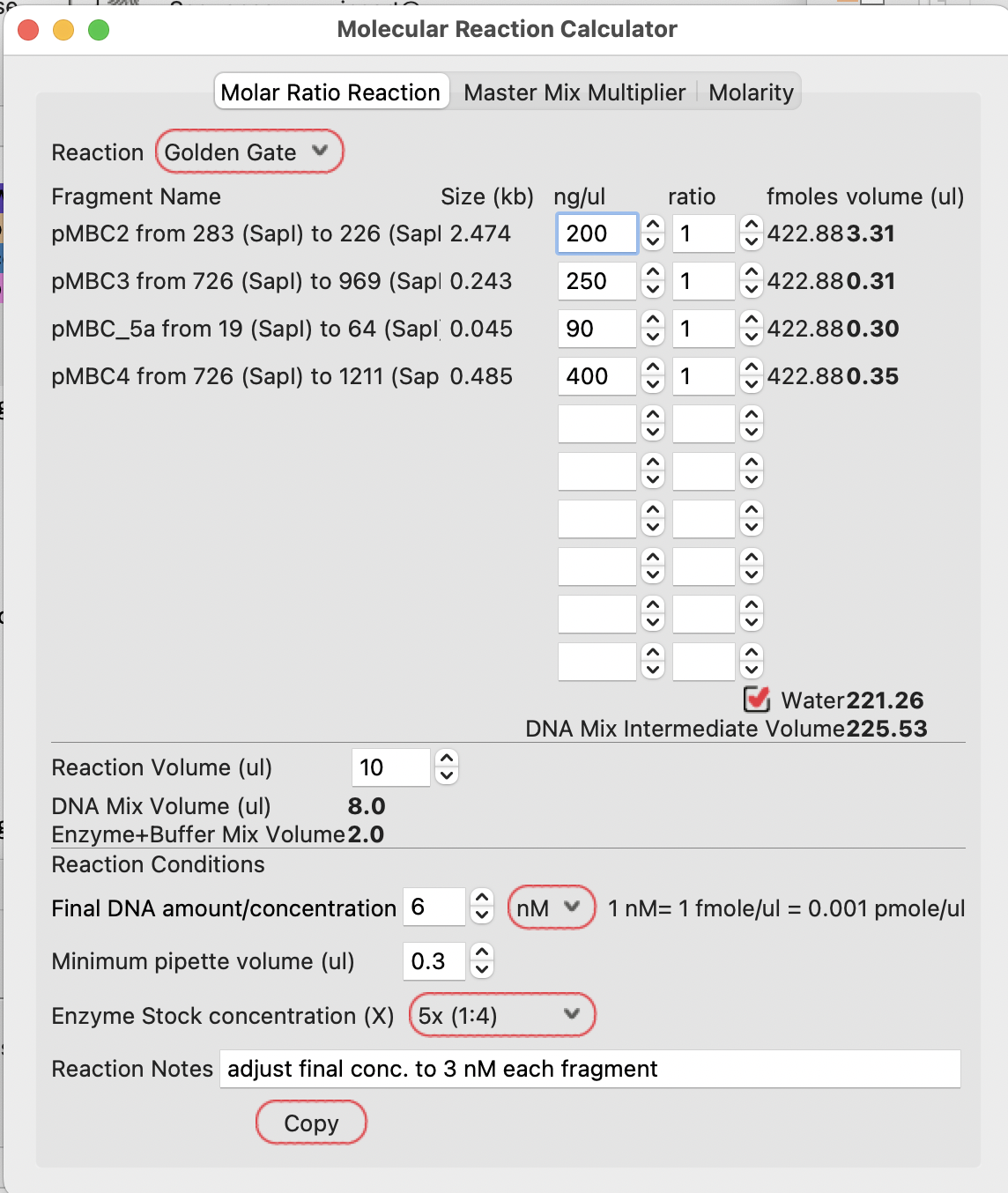
Given the plasmid concentrations, the calculator will show the volumes required to be mixed together to give the correct ratios of each DNA.
The case below calculates 0.58, 0.49, 0.53 and 0.30 ul of each DNA.
Note that the lowest volume is set to 0.3ul. If you want to calculate using lower volumes, you can set the minimum pipette volume to a lower value.
This Golden Gate recipe uses 3 nM each DNA final concentration in the reaction. With four fragments, the user should set the final DNA concentration to 12 nM (4 fragments at 3nM each).
The example below shows to add 17.51 ul of water to give the final required concentration of total DNA.
Because the Golden Gate reaction in this example is specified as a 5x enzyme stock concentration and a 10 ul reaction volume, the calculator shows using 8ul of DNA mix (from the 19.41 ul intermediate DNA mix) and 2 ul of enzyme mix in the final reaction.
These values can be adjusted as preferred by the user.